This mini post show a guide on how to use NCS Dummy to enable digital speed dispay in the instrument cluster.
The NCS Dummy makes it simple and safer to edit the trace files. Moreover, the NCS Dummy also allows users to assemble and disassemble NCS Expert data files to apply certain functionalities, which can't be done using the stock NCS Expert installation. Special thanks to Evan Goyuk for this awesome video guide. Check out more of his videos here. I have Wibbles, He's going to do a one to one with me and go over it, he told me to use NCS DUMMY profile instead. So i just did a little practice, managed to load my chasis and read the CAS, it gave me the 'strings', managed to get the ECU to read all the modules and did a backup of the FSWPSW.TRC. 4) Run NCS Dummy software 5) Select BMW Chassis and module, click magnifier button to open the Find window. Enter search item “SPEEDLOCK” and click “Find Next” (or click ENTER until you stumble upon the desired functions in the graphical trace editor: 'SPEEDLOCKXKMHMIN' and 'SPEEDLOCKXKMHMAX'.
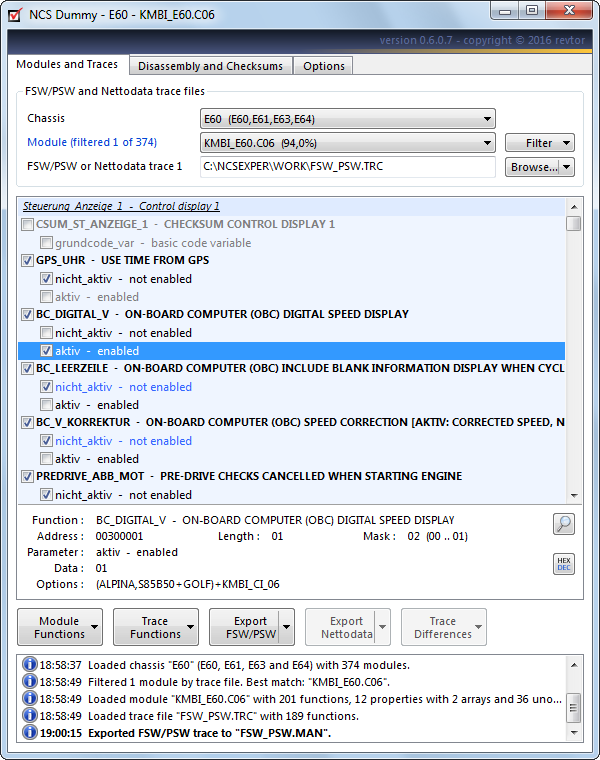
A frequently requested feature is a digital speed display in the instrument cluster. On certain BMW models like the E60 5 Series and E89 3 Series this just happens to very easy to enable. Function keyword “BC_DIGITAL_V” in the instrument cluster controls this feature.
Read the FSW_PSW.TRCtrace file from the instrument cluster with NCS Expert (see Reading FSW_PSW.TRC and NETTODAT.TRC trace files)and launch NCS Dummy.
1.Set the chassis.

NCS Expert (with English menus and buttons, and NCS Dummy Profile pre-installed) - NCS Dummy 4.0.1 - Tool32 4.0.3 - WinKFP 5.3.1 - Integrated SP-DATEN v53.3 - BMW Coding Tool v2.5.0 (for use as an alternative to NCS Dummy, or to update DATEN files as new ones become available). NCS Dummy is a tool that works with NCS Expert. It makes it easier to understand and make changes to the software config of your various ECUs. (Compared to using NCS Expert alone).
2.Set the module.
3.Load the trace file either by clicking the “Browse…” button, dragging the file to the textbox or,quicker, by choosing “Load FSW_PSW.TRC” from the button dropdown menu.
4.Go to function “BC_DIGITAL_V” in the graphical trace editor. You can make use of the powerful find functionality (CTRL+F or magnifier button) to quickly locate keywords in the trace file. The function has parameter “nicht_aktiv” checked which in this case means the digital speed display is disabled.Tick the checkbox of parameter “aktiv” to enable the feature.
5.Export your changes to the FSW_PSW.MAN trace manipulation file for NCS Expert: Click the “Export FSW/PSW” button and select “Export FSW_PSW.MAN” from the dropdown menu. If “Enable Quick Export” is ticked it suffices to simply click the “Export FSW/PSW” button to export the FSW_PSW.MAN trace manipulation file.
Finally code the FSW_PSW.MAN trace manipulation file back to the instrument cluster with NCS Expert (see Coding modules with FSW_PSW.MAN trace manipulation file) and you are done.A simple change like this could be done with a plain text editor as well, but NCS Dummy makes the process easier, quicker and safer. For more advanced changes a plain text editor no longer suffices and NCS Dummy is the only way to achieve your goals. In fact, NCS Dummy allows you to achieve things that are otherwise simply impossible with NCS Expert! A more advanced example is given in the next chapter.
How To Use Ncs Dummy Text
Readers who read this article also read:
How To Use Ncs Dummy Test
Ncs Expert For Dummies Download
- job_title = None Job title in PHENIX GUI, not used on command line
- input_files
- pdb_in = None Input PDB file
- ncs_in = None File with NCS operators to be applied to pdb_in This can be a .ncs_spec file from find_ncs, simple_ncs_from_pdb or find_ncs_from_density It can also be NCS formatted for resolve, including rotations, translations, and centers NOTE: NCS operators only apply to a limited region within the unit cell. The center of this region for molecule 1 (the one you enter with pdb_in) is specified by the NCS center for molecule 1. If your molecule in pdb_in is not near this, then it will be brought as close as possible using space-group symmetry (as defined in the CRYST1 record of pdb_in) before application of NCS. If your molecule is closer to the center for operator k, then it will instead be brought to that center and then transformed to all other locations
- map_in = None CCP4 or MRC-style map file (required if trim_overlapping=True). Map must cover the entire molecule including NCS copies.
- output_files
- pdb_out = apply_ncs.pdb Output PDB file
- cif_out = None Output CIF-formatted file
- log = apply_ncs.log Output log file
- params_out = apply_ncs_params.eff Parameters file to rerun apply_ncs
- directories
- temp_dir = 'temp_dir' Optional temporary work directory
- output_dir = ' Output directory where files are to be written
- gui_output_dir = None
- ncs
- trim_overlapping = False Trim pieces that overlap (including those overlapping by NCS). Requires a full-size (not boxed) map file (ccp4/mrc). Also requires resolution of the map.
- resolution = None Map resolution (for trim_overlapping)
- match_copy = None Force input model to be associated with NCS copy match_copy This can be useful if your molecule center is close to an NCS center other than the one it is associated with
- no_match_copy = None Do not allow input model to be associated with this NCS copy no_match_copy. This can be useful if your molecule center is close to an NCS center other than the one it is associated with
- apply_operators_directly = False You can apply the NCS matrices directly to an input model, without considering one of the NCS copies to be the identity for this model.
- max_copies = None You can limit the number of NCS copies to fewer than the actual number
- start_copy = None You can start the NCS copies with any available copy
- only_start_copy = None Just use start_copy and do not try to arrange chain names
- unique_only = None Just use unique part of pdb_in
- chain_names = None You can specify the chain names to generate
- used_chain_id_list = None You can specify the chain names to generate
- use_space_group_symmetry = True Normally apply_ncs uses space-group symmetry read from your input PDB file to find the appropriate operator. To disable this say use_space_group_symmetry=False. This is what you should do if you are using space group P1 as a dummy space group for example.
- control
- verbose = False Verbose output
- raise_sorry = False Raise sorry if problems
- debug = False Debugging output
- dry_run = False Just read in and check parameter names
- resolve_command_list = None You can supply any resolve command here NOTE: for command-line usage you need to enclose the whole set of commands in double quotes (") and each individual command in single quotes (') like this: resolve_command_list="'no_build' 'b_overall 23' "